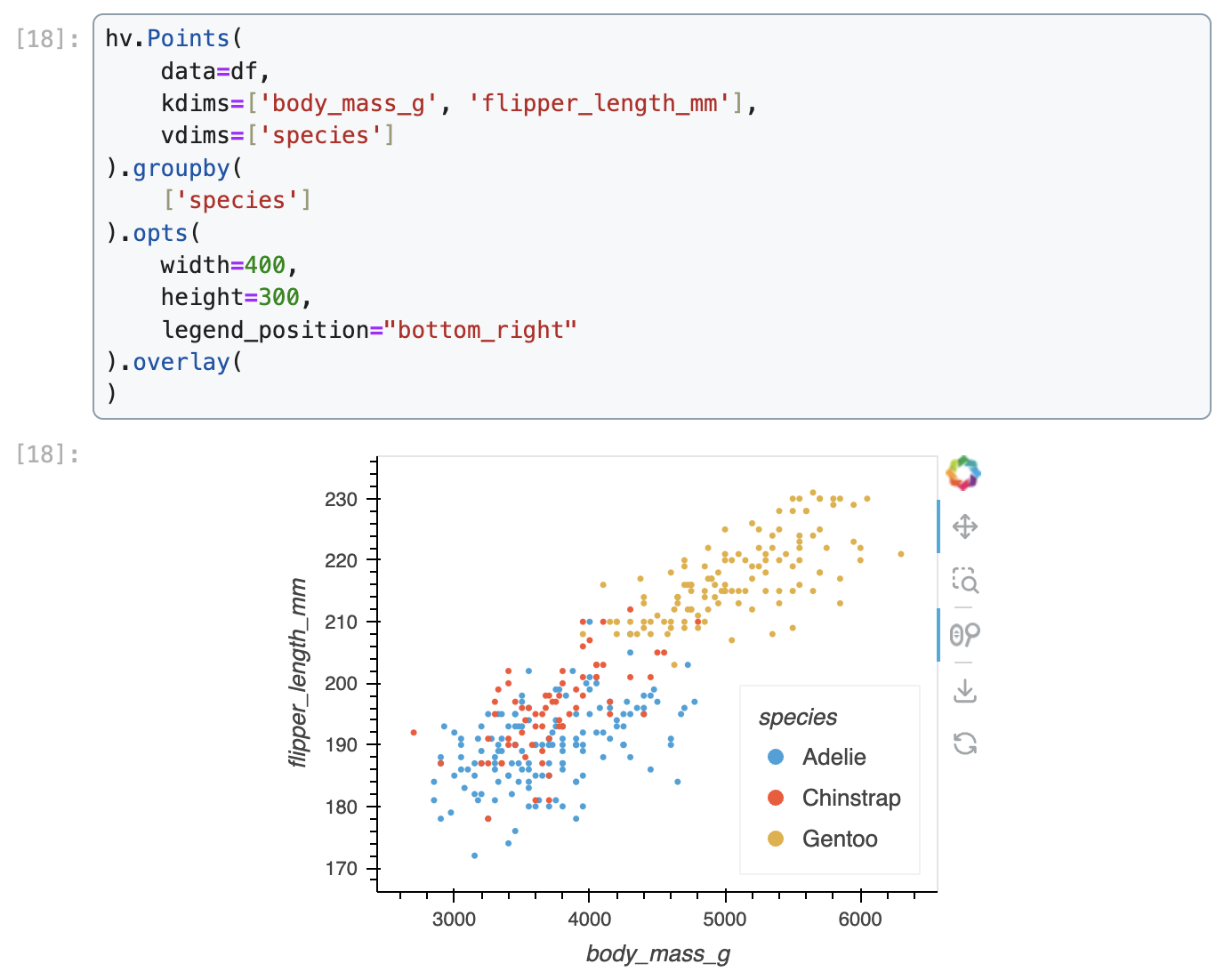
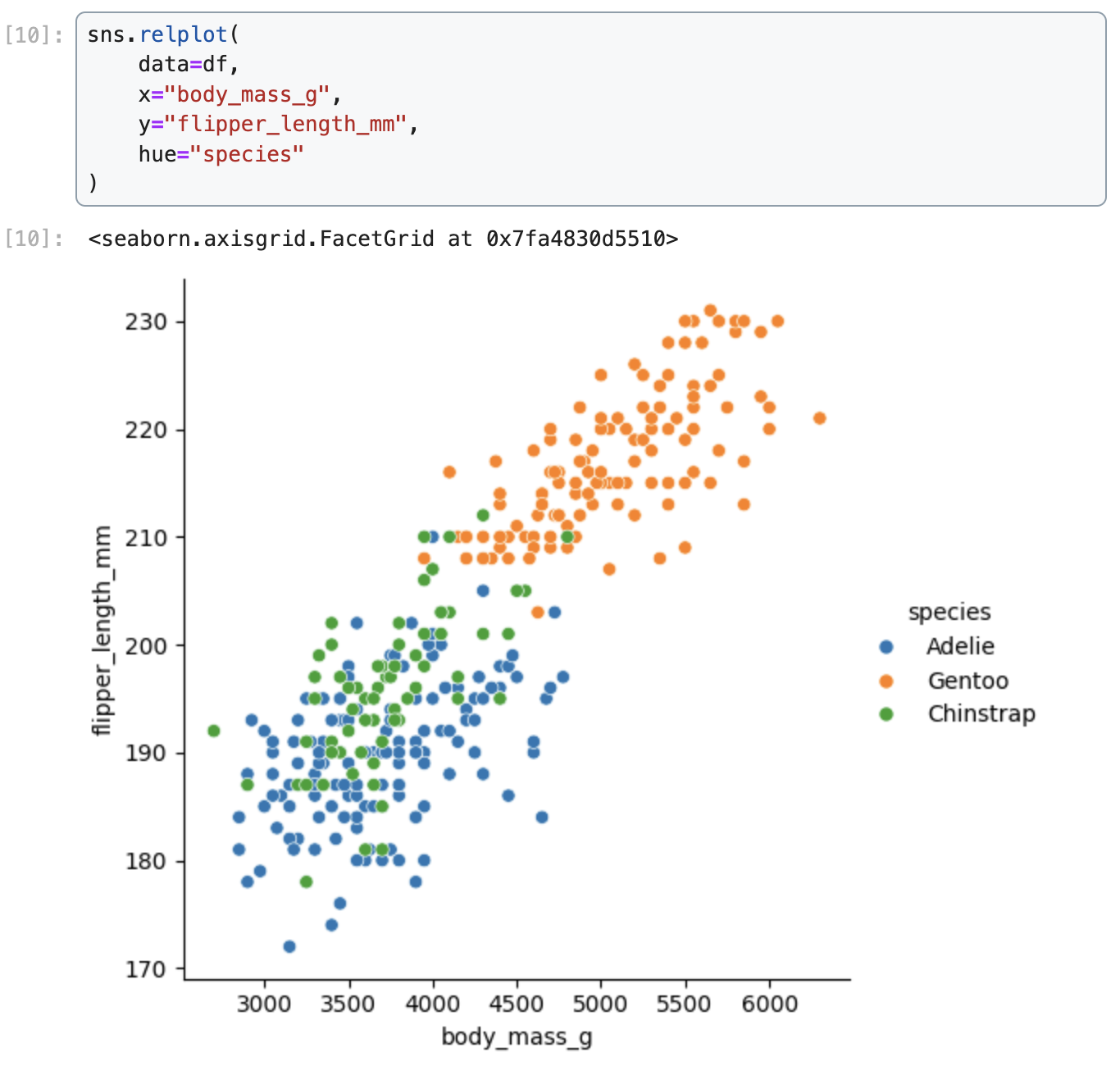
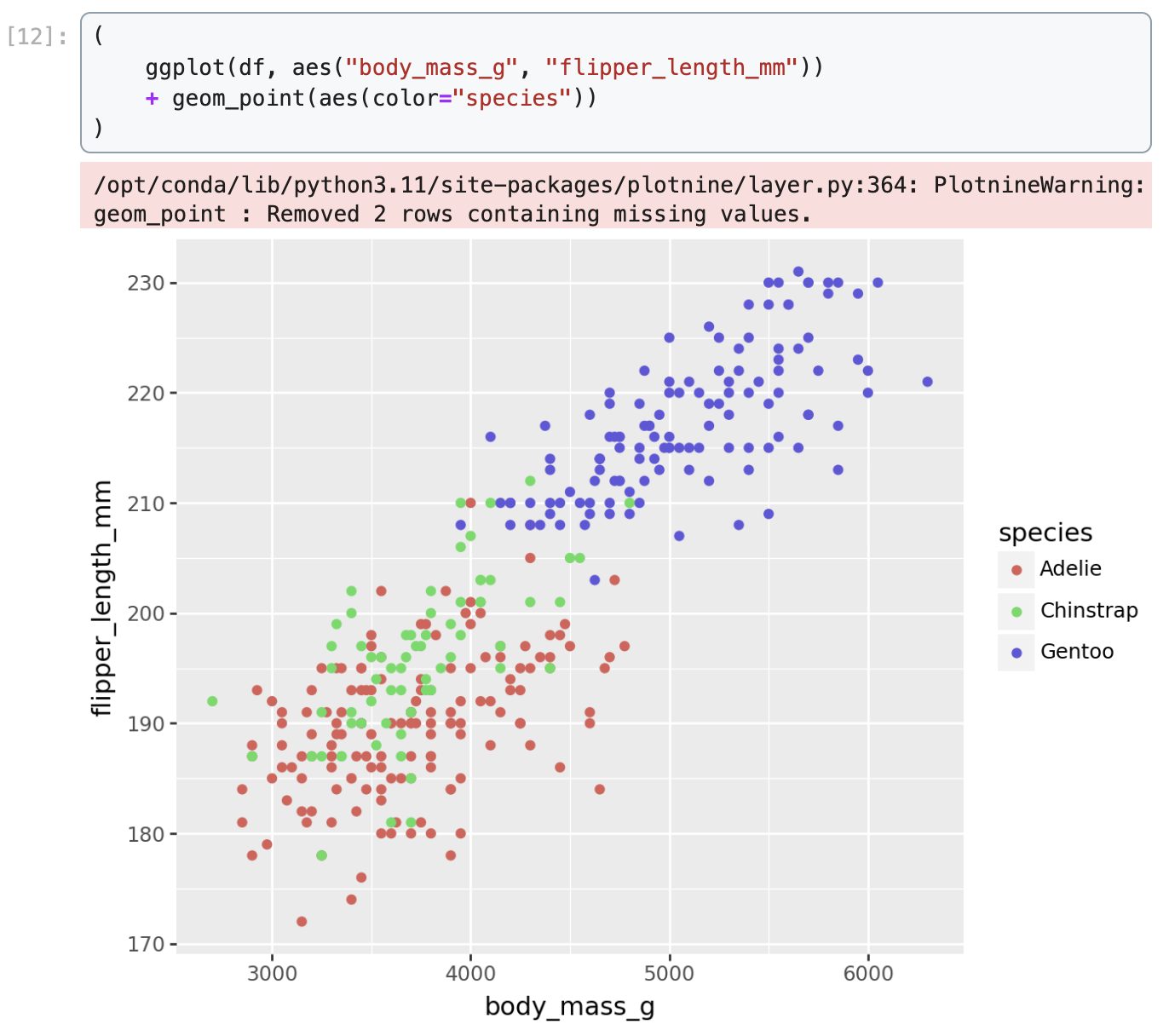
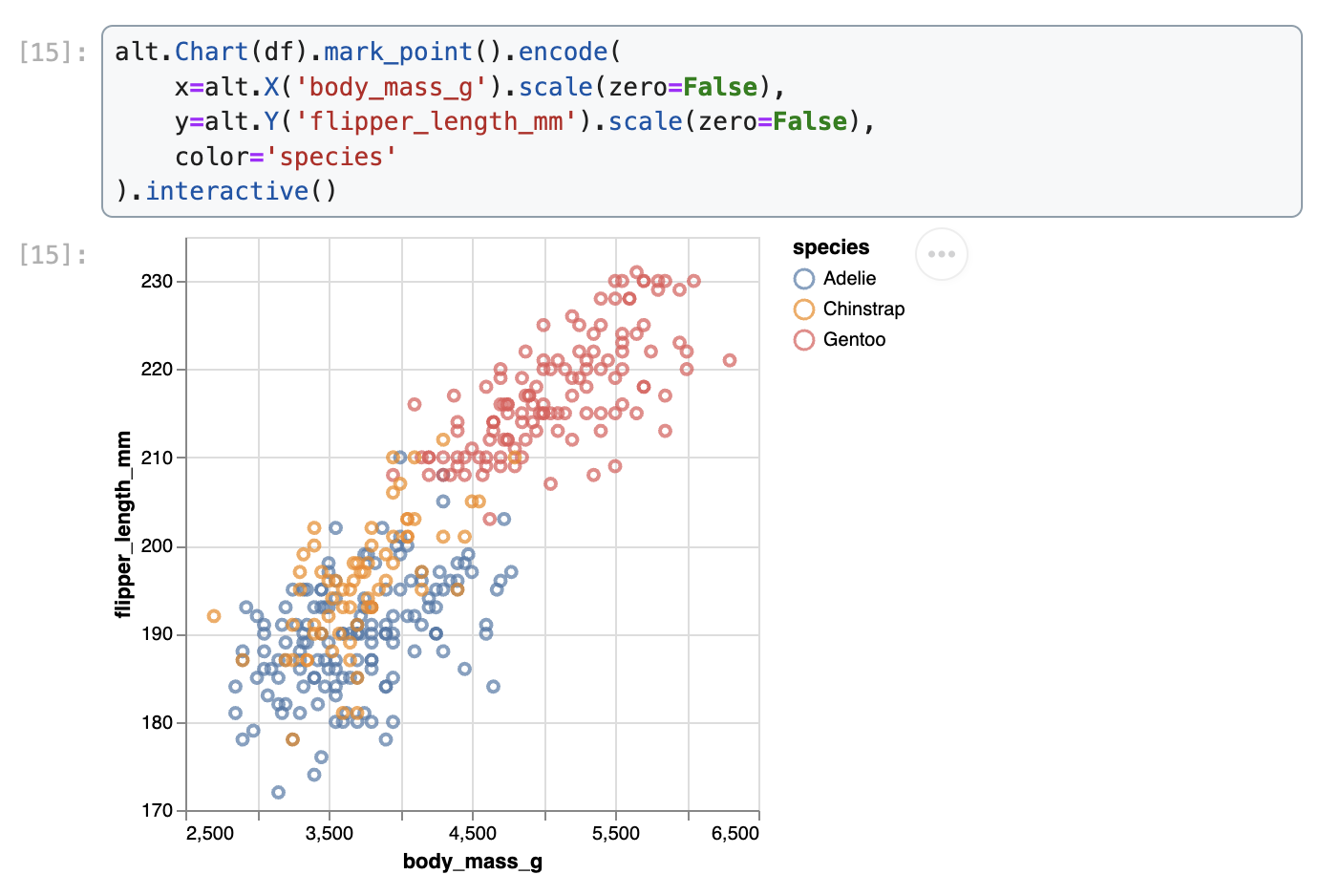
Data were collected and made available by Dr. Kristen Gorman and the Palmer Station, Antarctica LTER, a member of the Long Term Ecological Research Network.We demonstrate how to create a scatter plot of flipper length vs. body mass, colored by penguin species, in Matplotlib, Seaborn, Plotnine, Vega-Altair, and Holoviews.
Seaborn
Plotnine
Vega-Altair
Holoviews