Introduction
Sequencing runs with the Element AVITI output base calls and quality scores in bases files. The first step in analyzing the results of your AVITI sequencing run is to convert the bases files to FASTQ files through the Element Biosciences Bases2Fastq pipeline. We have adapted the Bases2Fastq pipeline to read and write data to your Mantle Database.Managing AVITI sequencing run data
In your Mantle Database, data is stored as entities, which have properties that can be files or metadata. AVITI sequencing run data can be stored in Mantle as entities of theelement_bases data type. Expand to view the properties of element_bases entities:
Properties
Properties
Directory containing outputs from an AVITI sequencing run
Used as an input to Bases2Fastq. Optional CSV file that specifies demultiplexing settings, FASTQ file settings, and sample information. If not provided, the default output run manifest from the run directory is used. See the Bases2Fastq documentation for more information.
Converting bases files to FASTQ files
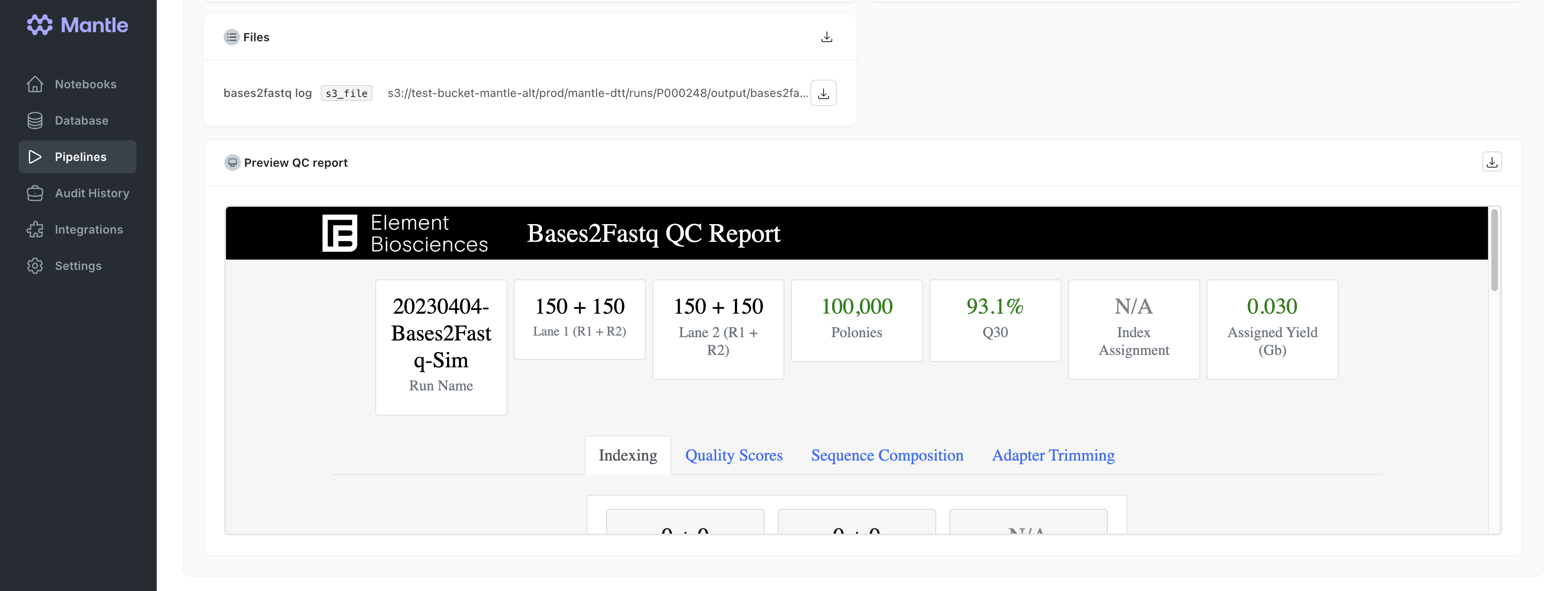
bases2fastq pipeline adapts the Element Biosciences Bases2Fastq pipeline to allow it to take a Mantle element_bases entity as input and to create a element_fastqs_directory entity with the results directory. Additionally, when you run the Mantle bases2fastq pipeline, the QC report is displayed on the pipeline run page. All pipeline results files can also be browsed on the pipeline run page. Learn more about the pipeline in the Element Biosciences Bases2Fastq documentation.